- info@nouvahub.com
- +977 58 528 0 538
Researchers sequenced the viral genome from samples in the state of Maharashtra and found a unique combination of three mutations that suggest the SARS-CoV-2 virus is continually evolving to evade the human immune response.
With the COVID-19 pandemic continuing to spread in several parts of the world, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is evolving to evade our immune system. Several new variants, designated variants of concern, have been reported from the United Kingdom, South Africa, and Brazil at the end of 2020, which seem to be more infectious than the original strain.
The introduction of new variants and their consequences is important in guiding public health response in a country. With the tremendous spike in COVID-19 seen in India, especially in the state of Maharashtra, the government increased the sequencing of the viral genome to identify possible new mutants and understand the fitness of new strains.
Researchers from several Indian institutes involved in SARS-CoV-2 research obtained samples from international travelers to Maharashtra. About 5% of the surveillance samples tested positive and were sequenced, and the whole genomes of SARS-CoV-2 analyzed.
The team also analyzed the crystal structure of the top 10 virus spike protein mutations complexed with the human receptor, angiotensin-converting enzyme 2 (ACE2). They also assessed the effect of mutations on binding to two neutralizing antibodies using the structures.
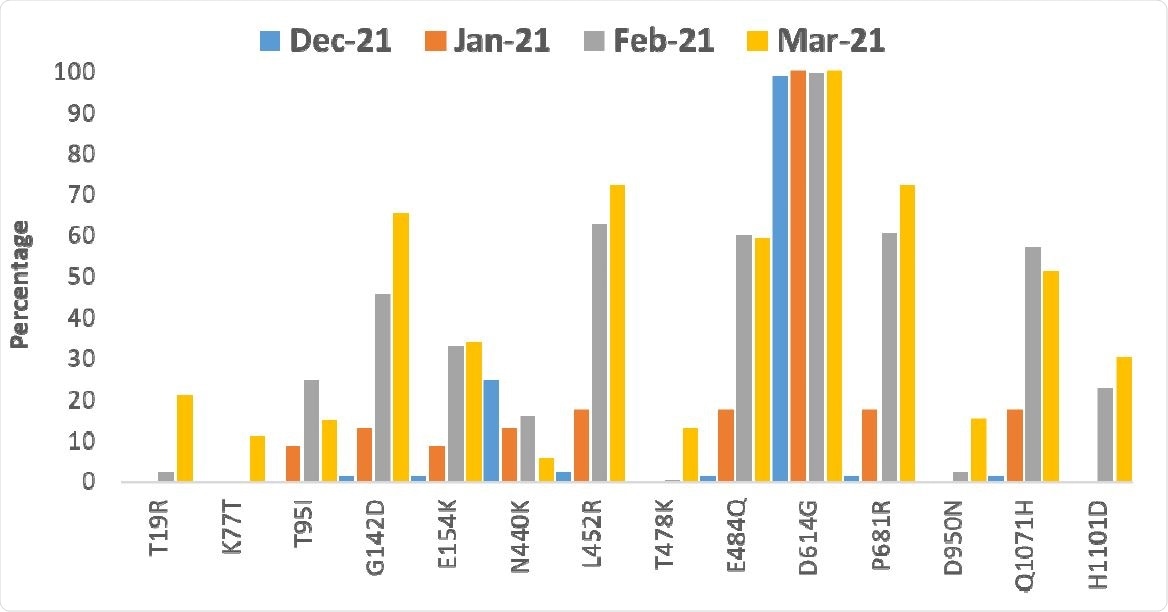
After analyzing 598 whole viral genomes, the team found 47 G lineages. The most common was the B.1.617, which was found in about half the genomes sequenced. Within the B.1.617 clade, they found four clusters linked to specific spike protein mutations.
They found that mutations L452R and E484Q in the receptor-binding domain (RBD) of the spike protein and the mutations G142D and P681R outside the RBD increased in frequency from January 2021. Other mutations included H1101D and T95I. Only a small proportion of the sequences showed the B.1.1.7 variant, particularly in December 2020.
One cluster of mutations did not include E484Q and had T19R and D950N mutations in the spike protein. Another mutation D111D, occurred along with the RBD mutations L452R and E484Q but was not seen in the cluster without the E484Q mutation. Analysis indicated an increase in the frequency of non-synonymous mutations (those that change protein sequences) since February 2021.
In western Maharashtra, places such as Mumbai, Pune, Thane, and Nashik showed several lineages apart from the dominant B.1.617 seen in the eastern part of the state.
1 Comment
A WordPress Commenter
May 1, 2021Hi, this is a comment.
To get started with moderating, editing, and deleting comments, please visit the Comments screen in the dashboard.
Commenter avatars come from Gravatar.